Challenge Overview
Important Links
- Submission-Review You can find your submissions artifacts here. Artifacts will contain output.txt's for both example test cases and provisional test cases with stdout/stderr and individual test case scores.
- Other Details For other details like Processing Server Specifications, Submission Queue, Example Test Cases Execution etc
Problem statement
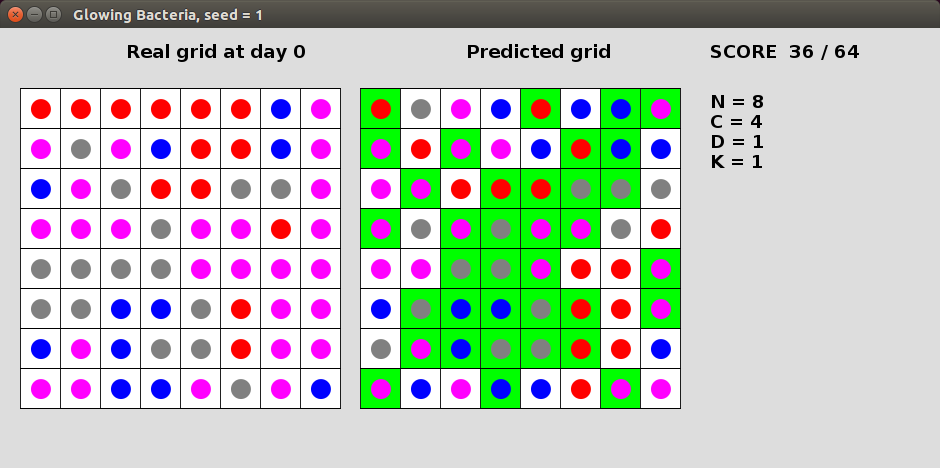
A group of glowing bacteria enjoy living on a NxN grid. These peculiar bacteria come in C different colours and change their colour every day based on the colour of their neighbours. In particular, a bacterium will take the colour of the minority (least frequent) colour of its neighbours that are between 1 and K (inclusive) squared Euclidean distance away from it. If there is no clear minority then the bacterium will keep its original colour. You are given a grid representing the bacteria colours after D days and your task is to predict their original colours on day 0. You will receive a point for each bacterium colour correctly predicted.Here is a solution for seed=1 with N=8, C=4, D=1 and K=1. The left grid shows the bacteria on day 0, while the right grid shows the bacteria after 1 day. In this case, each bacterium changes colour based only on its 4 directly adjacent neighbours (fewer at edges). In this solution we simply predict the same colour as in the input grid. The cells that have been predicted correctly are highlighted in green. This solution obtains a raw score of 36 out of a possible of 64 points.
Implementation
Your code will receive as input the following values:- N, the size of the grid.
- C, the number of bacteria colours.
- D, the number of simulation days.
- K, the distance threshold.
- NxN single-character lines describing the input grid, where line r*N+c represents the colour of the bacterium in row r and column c. These will be integers between 0 and (C-1), inclusive.
- On the first line, NxN - the number of cells in the grid.
- NxN single-character lines describing your predicted grid at day 0, where line r*N+c represents the predicted colour of the bacterium in row r and column c. These must be integers between 0 and (C-1), inclusive.
Scoring
Your raw score for a test case is the total number of correctly predicted bacteria colours. If your return was invalid (invalid return length, invalid characters or invalid colour ids etc.) then your raw score on this test case will be -1.If your raw score for a test case is negative then your normalized score for that test case is 0. Otherwise, your normalized score for each test case is YOUR/MAX, where YOUR is your raw score and MAX is the largest positive raw score currently obtained on this test case (considering only the last submission from each competitor). Finally, the sum of all your test scores is normalised to 100.
Test case generation
Please look at the visualizer's source code for the exact details about test case generation. Each test case is generated as follows:- The grid size N is randomly chosen between 8 and 50, inclusive.
- The number of colours C is randomly chosen between 2 and 8, inclusive.
- The number of simulation days D is randomly chosen between 1 and 5, inclusive.
- The threshold distance K is randomly chosen between 1 and 10, inclusive.
- Randomly generate the initial grid as follows:
- 1. Generate a random location (r, c) inside the grid.
- 2. Generate width W and height H, both randomly chosen between 1 and N/4 (integer division), inclusive.
- 3. Fill a rectangle with a top-left corner at (r, c), width W and height H with one of C colours chosen at random.
- 4. If there are still cells left uncoloured then repeat from step 1, otherwise terminate.
- Simulate D days to produce the input grid.
- All generated values are chosen uniformly at random.
Notes
- The squared Euclidean distance between cells (r1, c1) and (r2, c2) is (r1-r2)*(r1-r2) + (c1-c2)*(c1-c2).
- The time limit is 10 seconds per test case (this includes only the time spent in your code). The memory limit is 1024 megabytes.
- There is no explicit code size limit. The implicit source code size limit is around 1 MB (it is not advisable to submit codes of size close to that or larger). Once your code is compiled, the binary size should not exceed 1 MB.
- The compilation time limit is 30 seconds.
- There are 10 example test cases and 100 full submissions (provisional) test cases. There will be 2000 test cases in the final testing.
- The match is rated.
Languages Supported
C#, Java, C++ and Python
Submission Format
Your submission must be a single ZIP file not larger than 500 MB, with your Source Code only:
Please Note: Please zip only the file. Do not put it inside a folder before zipping, you should directly zip the file.
- Java Source Code - GlowingBacteria.java
- C++ Source Code - GlowingBacteria.cpp
- Python3.6 Source Code - GlowingBacteria.py
- C# Source Code - GlowingBacteria.cs
Sample Submissions
Tools
Submission format and an offline tester are available below. You can use it to test/debug your solution locally. You can also check its source code for an exact implementation of test case generation and score calculation. You can also find links to useful information and sample solutions in several languages.DOWNLOADS
HELPFUL INFORMATION
- Marathons Cheatsheet (a collection of useful links)
- Topcoder Cookbook on GitHub
- Topcoder Cookbook forums
OFFLINE TESTER / VISUALIZER
In order to use the offline tester/visualizer tool for testing your solution locally, you'll have to modify your solution by adding the main method that interacts with the tester/visualizer via reading data from standard input and writing data to standard output.Here are example solutions for different languages, modified to be executed with the visualizer. They all implement the same approach: predict the same colours as in the input grid.
- GlowingBacteria.cpp
- GlowingBacteria.java
- GlowingBacteria.py
- GlowingBacteria.cs
- You may modify and submit these example solutions.
java -jar tester.jar -exec "<command>" -seed <seed>
Here, <command> is the command to execute your program, and <seed> is seed for test case generation. If your compiled solution is an executable file, the command will be the full path to it, for example, "C:\TopCoder\solution.exe" or "~/topcoder/solution". In case your compiled solution is to be run with the help of an interpreter, for example, if you program in Java, the command will be something like "java -cp C:\TopCoder Solution".
Additionally you can use the following options:- -seed <seed>. Sets the seed used for test case generation, default is seed 1.
- -novis. Turn off visualization.
- -debug. Print debug information.
- -size <cell size>. Use custom grid size, default is 15 pixels.